Our research
We explore how old cells are replaced during physiological conditions, and how cell replacement and lineage progression are affected by injuries like stroke, spinal cord injury or myocardial infarction, and by diseases like cancer or multiple sclerosis.
One driving goal of our lab is to help the body replace worn-out or damaged cells and tissues. To reach this goal, one approach is to recruit and boost the body’s own mechanisms for cell replacement. To achieve this, it is important to identify cells with the capability to replace others, and to understand how they are controlled. To this end, we have a keen interest in developing methods to interrogate cell renewal and cell lineage relationships in humans.
Cell generation and lineage tracing in mice and humans
Using retrospective birth dating to study the turnover of cells in humans
While some tissues, like skin and bone marrow, have a high cell turnover rate, other areas like the brain seem to have minimal cell replacement. There is limited knowledge about different cell populations’ turnover dynamics in the human body due to technical challenges in studying this process.
A retrospective birth dating strategy developed in our lab takes advantage of the integration of nuclear bomb test-derived carbon-14 in the DNA during cell division. Using carbon dating, we investigate the turnover of different cell types in the body. In the brain, we study neurons, oligodendrocytes and microglia. We study cell turnover both in the healthy brain and in brains from patients suffering from stroke, multiple sclerosis, Alzheimer’s disease, Parkinson’s disease and depression. Carbon dating has been used to study the turnover dynamics of cells outside the brain, like heart cells and T-cells.
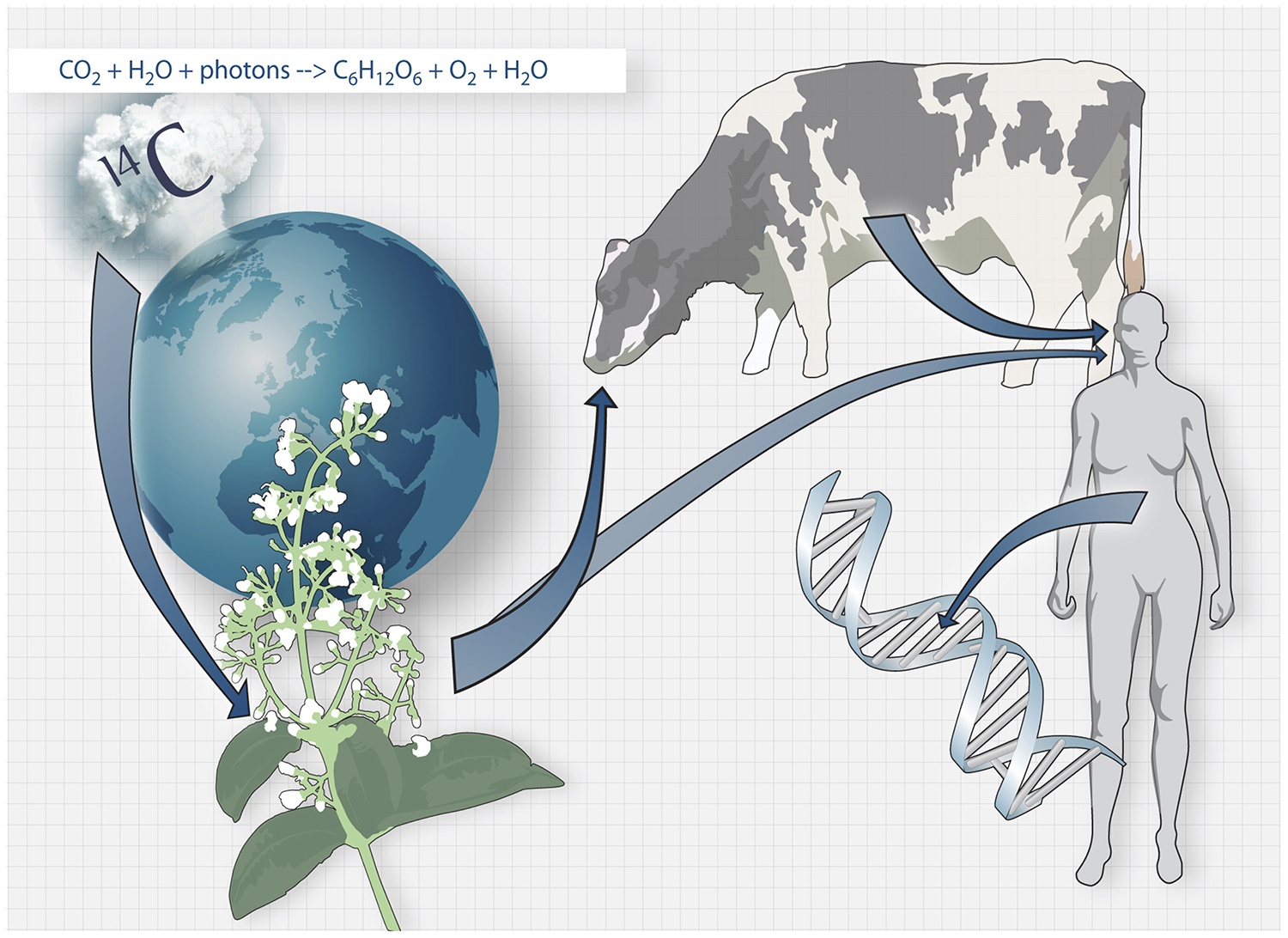
The carbon dating method. Carbon-14 generated by nuclear bomb tests during the Cold War caused the concentration of Carbon-14 in the atmosphere to increase drastically. Carbon-14 from the atmosphere is taken up by the biosphere through photosynthesis, and when we consume plants and animals that live off plants, the Carbon-14 is taken up by our bodies and integrates in our DNA during cell division.
Using somatic mutations to build family trees of cells in humans
All cells in our body descend from a single fertilized egg cell. The relationships between our cells can be thought of as a large family tree at the cellular level. Not much is known about the relationships between cells in humans, and knowledge of these relationships could shed light on the origins of new cells and the precise dynamics of cell turnover in humans.
Recent technological advances for single-cell DNA sequencing has opened up the possibility of tracing cell lineage in humans by analysis of somatic mutations acquired during cell divisions. Through analysis of somatic mutations, we study the origins and phylogenetic relationships of human cells. Within the scope of these projects, we are developing sample preparation protocols for single-cell DNA sequencing, as well as bioinformatic tools for analysis of single cell DNA sequencing data and somatic variant calling in single cells.
Researchers: Ilke Demirci, Joanna Hård, Jeff Mold, Marta Paterlini, Mathew Tata
Simultaneous profiling of cell types and lineages in the mouse brain
Tracing cell lineages in complex tissues, like the mammalian brain, involves marking a subset of cells and following their descendants. Traditional strategies like recombinase-mediated activation of fluorescent reporter gene expression in mice can be used to study cell lineages, but that requires sparse labelling of cells and does not permit large-scale reconstruction of lineages. Meanwhile, single-cell RNA sequencing allows large-scale characterization of molecular states and has revealed hundreds of distinct cell types in highly complex tissues such as the brain, but this method does not inform us about cellular relationships.
To overcome the limitations of both methods, we developed a high-throughput approach that allows simultaneous cell type identification and reconstruction of clonal relationships using single-cell RNA sequencing. Our approach involves the delivery of a highly diverse lentiviral library for genetic barcoding of neural stem cells in the developing mouse brain followed by RNA sequencing of labelled cells from the postnatal brain. We have sequenced the transcriptomes of thousands of individual cells from different anatomical regions and established the clonal relationships among the identified cell types yielding novel insights into the lineage potential of neural stem cells. The approach is widely applicable to simultaneously profile cell types and lineages in different tissues during development, homeostasis and disease.
Central nervous system injury and repair
Astrocytes as a reservoir for new neurons in the brain
In the brain, recruiting endogenous growth to boost cell replacement is perhaps more challenging than in many other organs because the brain’s own ability to replace neurons is almost non-existent. Neural stem cells in the adult brain are specialized astrocytes. Does that mean that, conversely, other astrocytes can be regarded as specialized neural stem cells? We previously found that some astrocytes, one of the most abundant cell types in the brain, harbor a latent capacity to generate neurons in vivo. Now, we are curious to understand this neurogenic capacity in more detail. One explicit goal of our research is to recruit parenchymal astrocytes as a reservoir for new neurons throughout the central nervous system.
Researchers: Giuseppe Santopolo, Margherita Zamboni
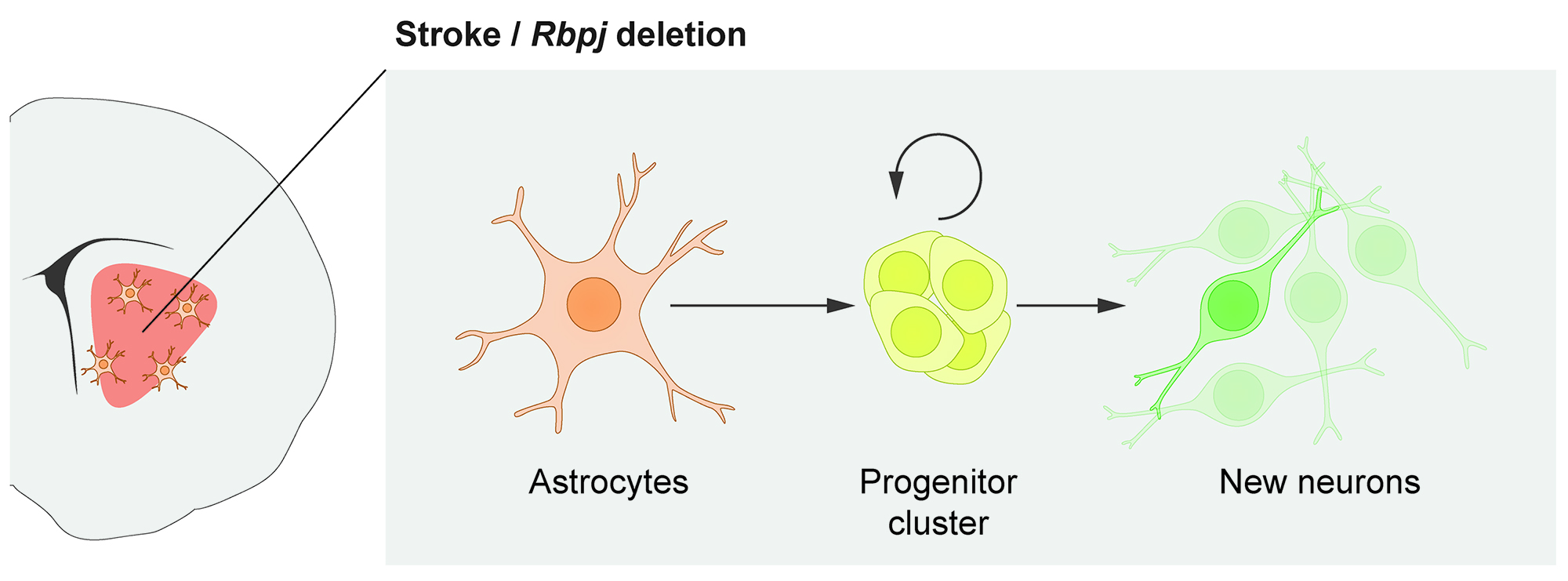
In response to stroke, some astrocytes in the mouse striatum activate a latent neurogenic program and generate neurons. This neurogenic capacity can be activated even in healthy mice by deleting the Notch-mediating transcription factor Rbpj.
Neural stem cells in the adult spinal cord
The neural stem cells in the adult spinal cord are ependymal cells. After spinal cord injury, they generate more than half of the astrocytes in the glial scar that forms at the lesion site, and a small fraction of remyelinating oligodendrocytes. We have shown that the ependymal cell contribution to the glial scar is essential to regain tissue integrity and limits the loss of neurons after spinal cord injury. These findings identify ependymal cells as an interesting target for manipulating the cellular response to spinal cord injury to preserve or regain function.
We are currently working on delineating the exact identity and properties of neural stem cells within the ependymal cell population in the spinal cord, as well as exploring the underlying mechanisms that control their reaction to injury.
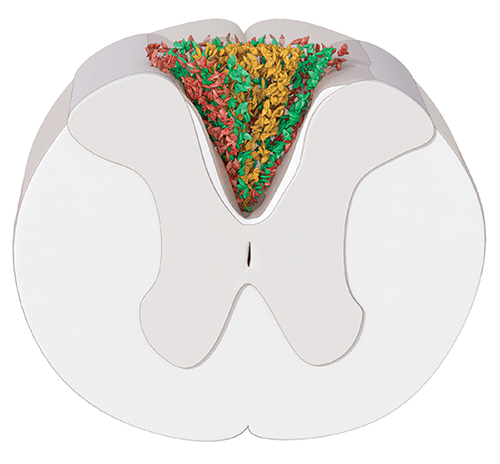
The scar formed after spinal cord injury contains astrocytes surrounding a fibrotic core (yellow). The inner layer of astrocytes (green) is derived from ependymal cells.
Cancer, immunology and cancer immunology
Clonal dynamics of the hematopoetic system
We are interested in the clonal dynamics of biological systems. We have investigated this primarily in the context of healthy human T cell populations because these cells can be tracked easily on the basis of their shared T cell receptors. In the future, we aim to extend these studies to other hematopoietic lineages in healthy and disease states and other organ systems. The long-term goal of these studies is to better understand how complex tissues are built and maintained at the level of individual cells and their progeny. This research could have implications for regenerative medicine as well as for determining how ageing and injuries lead to malignancies in different tissues.
Spatial analysis of tumor-infiltrating lymphocytes
In the past decade, immunotherapies, which mainly target T cells, have revolutionised anti-cancer care. In addition to T cells, data suggest that other immune cells, such as B/plasma cells, can also regulate tumor progression. Despite our growing knowledge of tumor-infiltrating lymphocytes, their clonal dynamics, particularly with respect to their spatial distribution, remains unclear. To address this knowledge gap, we are using existing, as well as developing new, single cell sequencing tools to uncover the transcriptional, clonal, and spatial distribution of lymphocytes in the tumor microenvironment.
The role of human cytomegalovirus infection in prostate cancer
Persistent viral infections can regulate cellular functions in health and disease. Human cytomegalovirus is a common herpes virus that inhabits cells of the hematopoietic lineage, but has also been proposed to inhabit the prostate gland. We study the characteristics of human cytomegalovirus infection in benign and malignant cells of the prostate, and explore human cytomegalovirus in prostate cancer as a potential therapeutic target.
